Ideogram Viewer

| Overview | |
| Description | An interactive tool for viewing the location and annotation of genes on human chromosomes |
| Development Information | |
| URL | https://bioinformatics.mdanderson.org/ideogramviewer/Ideogram.html?genelist1=ERBB2,TP53,BRCA1,PTEN,AGL&genelist2=TP53,MDM2,ERBB2,ERBB4&mirlist1=hsa-let-7a-2,hsa-let-7c,hsa-let-7e&mirlist2=hsa-let-7c,hsa-let-7a-2 |
| Language | JavaScript |
| Current version | 1.0.8 |
| Platforms | Web application |
| License | Not Required |
| Status | Active |
| Last updated | July 29, 2016 |
| Help and Support | |
| Contact | Bradley M. Broom |
Ideogram Viewer
Introduction
Ideograms provide a schematic representation of chromosomes. They are used to show the relative size of the chromosomes and their characteristic banding patterns.
We have developed an interactive viewer to show the location of the requested genes and microRNAs on human chromosomes. We collected a set of human oncogenes and tumor suppressor genes from different resources that can be chosen to display on chromosomes.
Viewer Features
- Displays the chromosome ideogram in a web browser
- Displays gene and/or microRNA markers and annotations for a list of gene symbols and a list of microRNA IDs
- Displays a subset of chromosomes
- Horizontal zooming and panning
- Links gene symbols and microRNA IDs annotation to different resources: NCBI Entrez, GeneCards, UCSC Genome Browser and geneSmash
- Selects a genomic region on a single chromosome and records the symbols to a new window
- Selects a genomic region on a single chromosome and opens this region in the UCSC Genome Browser
- Displays a set of oncogenes and suppressor genes
- Drag slide controls to view horizontal portion of the viewer
Feature Description
All chromosomes and cytobands can be viewed via Ideogram
To view specific genes and/or microRNAs, a user can specify query parameters “genelist1”, “genelist2” (a list of comma separated HGNC gene symbols), “mirlist1” and “mirlist2” (a list of comma separated microRNA IDs) to mark them on chromosomes and display annotations, for example genelist1=TP53&genelist2=ERBB4&mirlist1=hsa-let-7a-2&mirlist2=hsa-let-7c . A specification for gene symbols and labels is defined in the Legend color scheme below.
User can define a color scheme for genes and microRNA IDs by specifying query parameters “genelist1color”, “genelist2color”, “mirlist1color”, “mirlist2color”, “genelistmcolor”, “mirlistmcolor” (Both a color name such as ‘Blue’ or a HEX value ‘#FF0000’ (URL-encoding %23 for a ‘#’ sign are fine.) For example mirlist2color=black&genelist1color=%23ffff00 .
A query parameter “suppresslabel” can be used to optionally turn on/off label. For example suppresslabel=true ‘true’, ‘false’, ‘yes’, ‘no’, ‘1’, ‘0’ are valid values.
Mousing over a cytoband will display its annotation. Left mouse click a gene symbol or a microRNA ID to pop up a menu for viewing different annotation resources.
Display can be restricted to a subset of chromosomes by including a “chromosome” query parameter, for example chromosome=2,17,10 Chromosome numbers 1 to 22 and sex chromosomes X and Y are allowed values.
User can zoom in the diagram by using the mouse scroll wheel or double-clicking. User can zoom out the diagram by using the mouse scroll wheel or shift double-clicking. Alternatively, use the buttons at the top of the page, or use the control items at the end of the zoom indicator at the top of the ideogram.
Use the Reset button to restore the original zoom level.
User can pan the diagram by clicking and dragging across the Ideogram area. Alternatively, drag the context rectangle on the zoom indicator at the top of the ideogram.
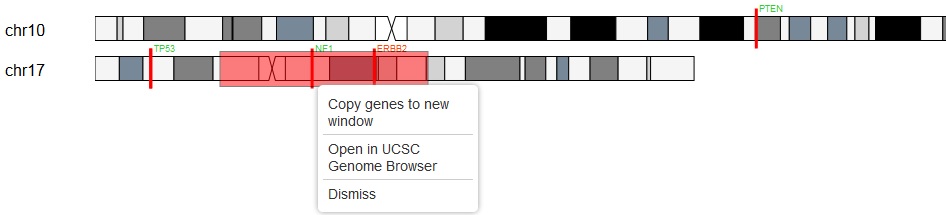
To select a region on a chromosome: Move the mouse cursor to a position on the chromosome, press the shift key, the cursor turns to cross hair style, left mouse down, move the cursor to select a region of your interest, release the mouse. If there are symbols inside the selected region, a menu allows user to copy the symbols to a new window or open the region in the UCSC Genome Browser.
User can click “oncogene” and/or “suppressor” legend items to display those cancer genes on the viewer with the current zoom level.
A slider at the top of the viewer indicates the viewer portion that is shown on the screen. User can drag the slider controls to view a horizontal portion of the viewer.
An example of displaying the gene markers and annotations:
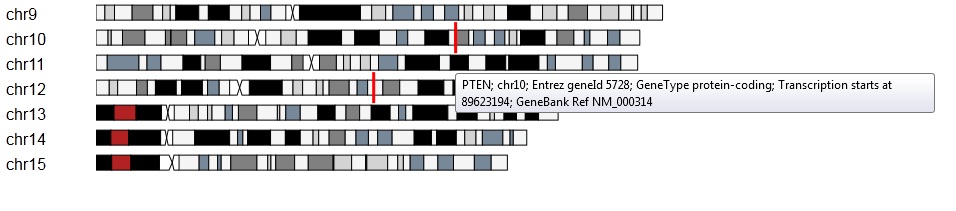
An example of selecting a genomic region on a chromosome:
Data sources
Cytoband information extracted from UCSC Genome Browser Download page is used to draw the Ideogram template. The data refer to February 2009 assembly of the human genome (hg19, GRCh 37 Genome Reference Consortium Human Reference 37).
Gene annotations are retrieved from geneSmash web service geneSmash
Lists of human oncogenes and tumor suppressor genes are collected from these resources: Cell Signaling , Wikipedia Oncogene , Wikipedia Suppressor gene and a publication from the journal Cancer Research Walker, et al. . (See its Supplementary Table ). We curated and compiled these cancer gene lists in house.
Cytoband color scheme
Each cytoband has a staining color and a banding pattern. Cytogenetic bands also provide potential predictions of the structural characteristics of a chromosome. Ideogram viewer uses cytoband colors consistent with Bioconductor’s R package, biovizBase: biovizBase .
Legend color scheme
The genes and microRNAs are indicated by the colored solid or dashed lines in the viewer.
Genes or microRNA Ids from query parameters in the website url:
- A gene from genelist is displayed as solid red line.
- A gene from genelist1 is displayed as a solid red line.
- A gene from genelist2 is displayed as a solid fuchsia line.
- A microRNA ID from mirlist1 is displayed as a solid deep blue line.
- A mircroRNA ID from mirlist2 is displayed as a solid sky blue line.
- A gene from both genelist1 and genelist2 is displayed as a dashed red line.
- A gene from both mirlist1 and mirlist2 is displayed as a dashed sky blue line.
Oncogenes and Suppressor genes from data files:
- Oncogenes are displayed as solid orange lines.
- Suppressor genes are displayed as solid lime green lines.
All labels for gene symbols or microRNA IDs by default are in black text. If a gene is from the parameter list and is an oncogene or a suppressor, the label is orange (for oncogene) or lime green (for suppressor).
Browser compatibility
This application will not work on all internet browsers. We have tested it on the following browsers:
- Firefox 25.0
- Safari 6.0
- Chrome 37.0
How to capture high quality images for publication
Please refer to this page How to capture high quality images for publication
Credits
Rong Yao, Chris Wakefield, Bradley Broom. We thank Kevin Coombes for creating the geneSmash web service for gene annotations. We thank the UCSC Genome Browser Project Team for providing the web resource to download cytoband data.