This project is archived and no longer maintained.
Bayes Mix User Guide
| Overview | |
| Description | A tool for performing model-based inference for differential gene expression, using a non-parametric Bayesian mixture probability model for the distribution of gene intensities under different conditions |
| Development Information | |
| Language | R and C |
| Current version | 0.8.8 |
| Platforms | Windows and Linux |
| Status | Inactive |
| Last updated | 15th August 2003 |
Bayes Mix User’s Guide
BayesMix is a model-based inference for differential gene expression, using a non-parametric Bayesian probability model for the distribution of gene intensities under different conditions, similar to the Empirical Bayes approach.
The BayesMix distribution consists of a Java front-end for an R back-end which calls various C functions implementing the statistical method. A pseudoterminal application qua agent accepts commands from the Java front-end and returns output from R.
User Interface
The Java application processes matrix file data. The information input by the user is checked for validity. Invalid input will cause the offending field to be displayed in reverse video. ToolTips with numeric range information are provided. The display button will be enabled when all fields are considered valid.
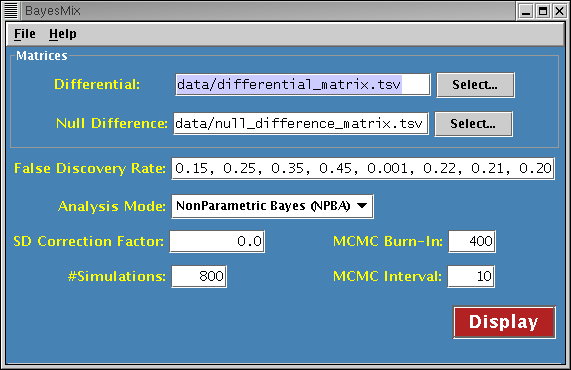
Differential Matrix
Specifies the filename of the differential matrix. The matrix file must have its fields separated by tabs. The first line of the file [header] contains the names of the variables (columns), followed by some number of lines containing the data to be analyzed. Each data line begins with a row name, followed by data values. The header line should contain one less column than a data line. It is recommended that all header fields be quoted to avoid ambiguity.
Null Difference Matrix
Specifies the filename of the null difference matrix. The format of the matrix is the same as that of the differential matrix.
False Discovery Rate
Specifies a list of real numbers representing false discovery rates. There can be a minumum of four and a maximum of ten values. Each value must be within the range [0.0, 1.0).
Analysis Mode
Specifies the desired processing to perform. Choices are:
- NonParametric Bayes (NPBA)
- Empirical Bayes (EBA)
- Sequential EBA-NPBA
SD Correction Factor
Specifies the calibration factor(s) for standard deviation correction. There can be either one or two real number values. When two values are provided, the first is used with the differential matrix and the second with the null difference matrix. If only one value is provided, it is used for both matrices. NPBA analysis mode only.
Simulations
Specifies a positive integer value representing the number of MCMC simulations to perform. NPBA or Sequential analysis mode only.
MCMC Burn-In
Specifies a non-negative integer value representing the number of MCMC simulations to discard as part of the burn-in. This value can be at most one half of the number of simulations. NPBA or Sequential analysis mode only.
MCMC Interval
Specifies a non-negative integer value representing the MCMC sample rate. As such, higher values will cause less samples to be gathered. At least ten samples are required to get meaningful output. NPBA or Sequential analysis mode only.
Menus
File Pulldown Menu
The File pulldown menu contains all the generic file handling options.
- Open…: Reads settings from a configuration file.
- Save…: Write current settings to a configuration file.
- Quit: Quits the application.
Help Pulldown Menu
The Help pulldown menu contains all the options providing basic assistance in using the application.
- Overview: Provides a high level description of the application’s purpose.
- User Guide: Displays this document via web browser.
- About: Provides information about the application itself.
Input/Output
Environment variables
- BMHOME: Root directory of the installation. Mandatory.
- BMCONF: Default directory for settings files. Defaults to $BMHOME/conf directory if unset.
- BMDATA: Default directory for data files. Defaults to $BMHOME/data directory if unset.
- BMOUTPUT: Directory for saving simulation output files. Defaults to $BMHOME/output directory if unset.
Files
- <settings>.cf: Configuration file used to store simulation settings.
- <output>.ps: Postscript file used to save graphs/plots.
- <datafile>.tsv: ASCII file that contains tab-separated values.
Command line activation
java
- Dbayesmix.home=pathname Specifies same thing as $BMHOME
- Dbayesmix.conf=pathname Specifies same thing as $BMCONF
- Dbayesmix.data=pathname Specifies same thing as $BMDATA
- Dbayesmix.output=pathname Specifies same thing as $BMOUTPUT
- Dnative.subdir=pathname Specifies a string in the format $MACHTYPE-$VENDOR-$OSTYPE
- Dstatistics.package.exec=r_filename Specifies the name of the R executable
- Dstatistics.package.args=r_exec_args Specifies arguments to be passed to the R executable
- Dmonitor.output=true Specifies output from the R executable should be echoed to the screen
- Djava.security.manager=default Enables the Java security manager
- Djava.security.policy=lib/security/BayesMix.policy Specifies the Java security policy file
- jar lib/bayesmix.jar Specifies the GUI application archive
- [ config_filename ] Specifies an existing BayesMix configuration file to open on startup