TCGA SpliceSeq
| Overview | |
| Description | A tool for investigating alternative mRNA splicing in TCGA tumor and adjacent normal samples |
| Development Information | |
| GitHub | MD-Anderson-Bioinformatics/SpliceSeq |
| URL | https://bioinformatics.mdanderson.org/TCGASpliceSeq/ |
| Language | HTML, Javascript, MySQL |
| Current version | 2.0 |
| Platforms | Platform independent |
| License | Freely available for academic and commercial use. |
| Status | Active |
| Last updated | August, 2016 |
| References | |
| Citation | Ryan M, Wong WC, Brown R, Akbani R, Su X, Broom B, Melott J, Weinstein J, TCGA SpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res (Databases Issue) 4 (D1) pD1018 (2016). https://doi.org/10.1093/nar/gkv1288 |
| Help and Support | |
| Contact | Insilico |
| Discussion | Issues On GitHub |
TCGA SpliceSeq
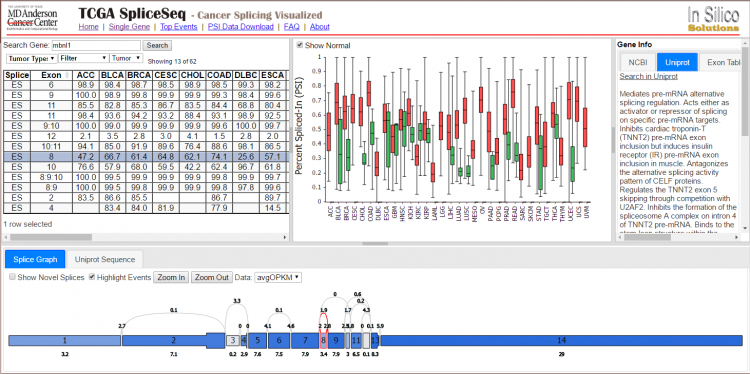
TCGA SpliceSeq is a web-based resource that provides a quick, user-friendly, highly visual resource for exploring the alternative splicing patterns of TCGA tumors. Percent Spliced In (PSI) values for splice events on samples from 33 different tumor types, including available adjacent normal samples, have been loaded into TCGA SpliceSeq. Investigators can interrogate genes of interest, search for the genes that show the strongest variation between or among selected tumor types, or explore splicing pattern changes between tumor and adjacent normal samples. The interface presents intuitive graphical representations of splicing patterns, read counts, and various statistical summaries, including percent spliced in. Splicing data can also be downloaded for inclusion in integrative analyses.