SpliceSeq
| Overview | |
| Description | A tool for investigating alternative mRNA splicing in next generation mRNA sequence data |
| Development Information | |
| Language | Java |
| Current version | 2.1 |
| Platforms | Platform independent |
| License | Freely available for academic and commercial use |
| Status | Active |
| Last updated | February 21, 2014 |
| References | |
| Citation | Ryan, M. C., Cleland, J., Kim, R.G., Wong, W. C., Weinstein, J. N., SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts, Bioinformatics 28 (18) p.2385 (2012). https://doi.org/10.1093/bioinformatics/bts452 |
| Help and Support | |
| Contact | MDACC-Bioinfo-IT-Admin@mdanderson.org |
Sample View
The Sample View is used to study alternative splicing patterns and their potential functional impact in single samples. To use this view, the sample must have already been loaded into the database. (See Data Loading for information on loading new samples)
To get to this view, first select Sample View in the View Combobox in the Data Selection Bar. Next, select the sample that you wish to investigate using the DataSet Combobox also in the Data Selection Bar.
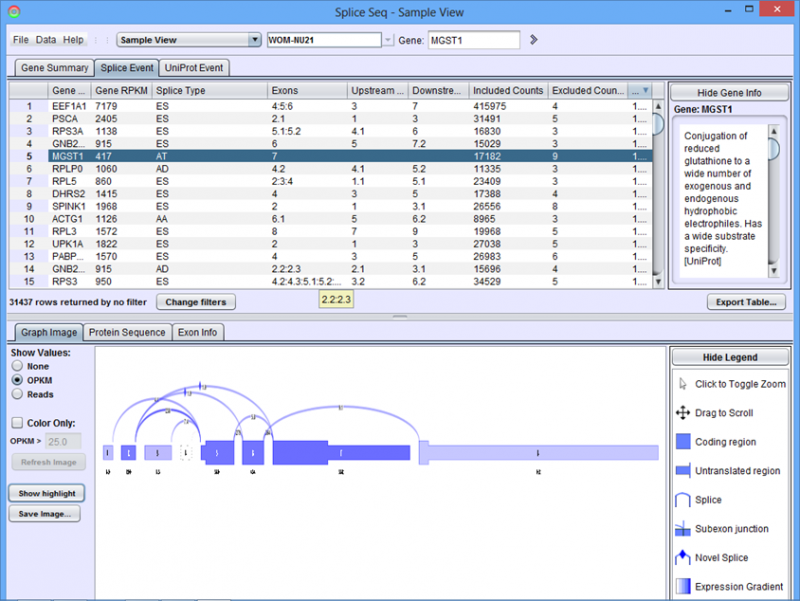
When the sample loads, the Results Panel will be loaded with all the splice events identified in the given sample. For a single sample, a splice event occurs when multiple isoforms of the transcript are expressed in the sample (e.g. some some transcripts include exon 10 and others splice it out)
Sample View Results Panel
Gene Summary Tab in the Results Panel will contain a row for every gene that is expressed in the sample.
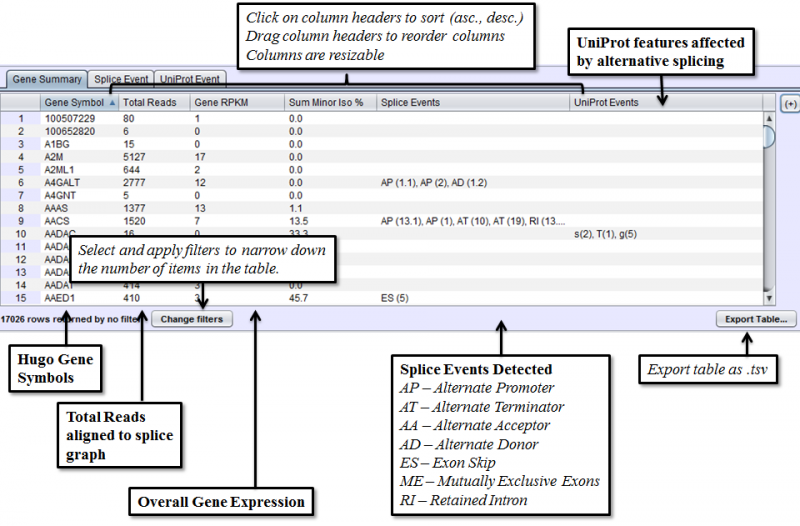
Clicking on a gene row will populate the Detail Panel with a splice graph representing the exons and splices that were present in the sample. When a gene is selected, the Gene Information Box will show a functional annotation of the selected gene from UniProt and/or NCBI Gene.
Columns that require further explanation are:
- Gene RPKM - Reads Per Kilobase per Million reads. RPKM is the total number of reads that uniquely aligned to the gene. It is normalized for gene length and total number of aligned reads in the sample. Gene RPKM provides an indication of coverage and expression level.
- Sum Minor Isoform % - Percentage of predicted mRNA expression that is not part of the major isoform. This can be used to find genes where alternative isoforms constitute more than a trivial percentage of gene expression.
- Splice Events - Categorized list of the splice events detected. For example, ES(4) indicates and exon skip on exon number 4. Use the Splice Event Filter drop down list to see the description and codes for each event type.
- UniProt Event - Categorized list of UniProt annotated regions that are affected by alternate splicing. For example, B(9) indicates a Bind Site on exon 9 that is excluded from some isoforms. See the legend in the Protein Sequence Tab of the Gene Panel for a full list of UniProt annotation categories
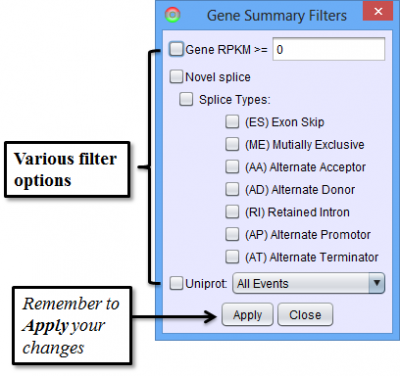
The items in the Results Panel can be filtered to find just the type of results that are of interest. To access filtering functions for the table, use the Change filters button. The following filters are available:
Splice Event Tab in the Results Panel will contain a row for every splice event detected.
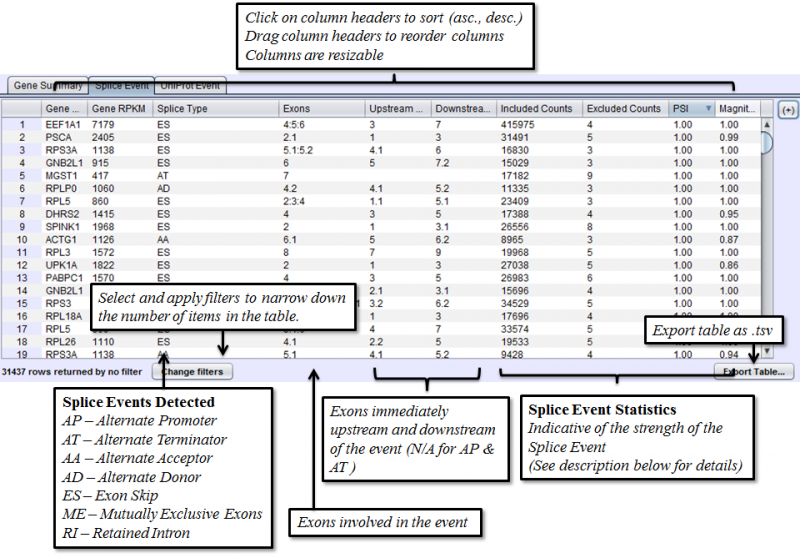
Columns that require additional explanation:
- Included Counts: The total number of reads on the included path (depends on the [Splice Event]https://bioinformatics.mdanderson.org/public-software/spliceseq/methods/#splice-event-detection)
- Excluded Counts: The total number of reads on the excluded path (depends on the [Splice Event]https://bioinformatics.mdanderson.org/public-software/spliceseq/methods/#splice-event-detection)
- PSI - Percent-Spliced-In. Percentage of the isoforms with the included path out of all the isoforms that express the given splice event
- Magnitude - The proportion of isoforms which the splice event affects
To access filtering functions for the table, use the Change filters button. The following filters are available:

Uniprot Event Tab in the Results Panel will contain a row for every UniProt annotated feature that is affected by alternative splicing.
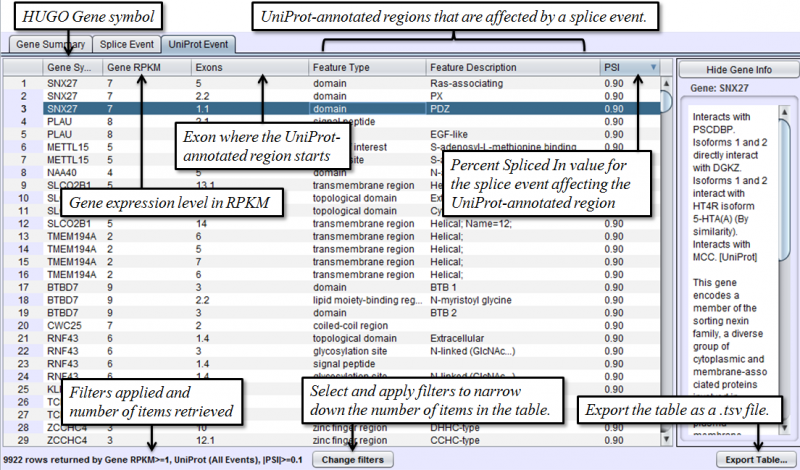
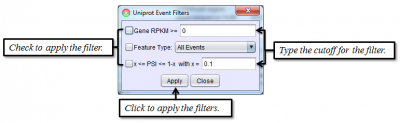
To access filtering functions for the table, use the Change filters button. The following filters are available:
The Show Values radio buttons on the left toggles the display of numerical values for every exon and splice. There are 3 options:
- None - No numerical values are displayed
- Reads - Total reads that included the exon / splice junction
- OPKM - Observations Per Kilobase per Million reads. OPKM is read total of the exon or splice normalized by length and total number of aligned reads in the sample. The length used is the window size in which a read may overlap the exon / splice. This normalization allows for cross exon or cross sample comparisons.
Protein Sequence Tab of the Detail Panel shows the protein sequence, functional annotation, and relative expression level of the isoforms inferred from splice patterns of the gene from the sample. For basic information on this tab, refer to Protein Sequence Tab.
The below example shows that differential expression of exons 1 and 2 results in different expression of the UniProt feature region CAP.
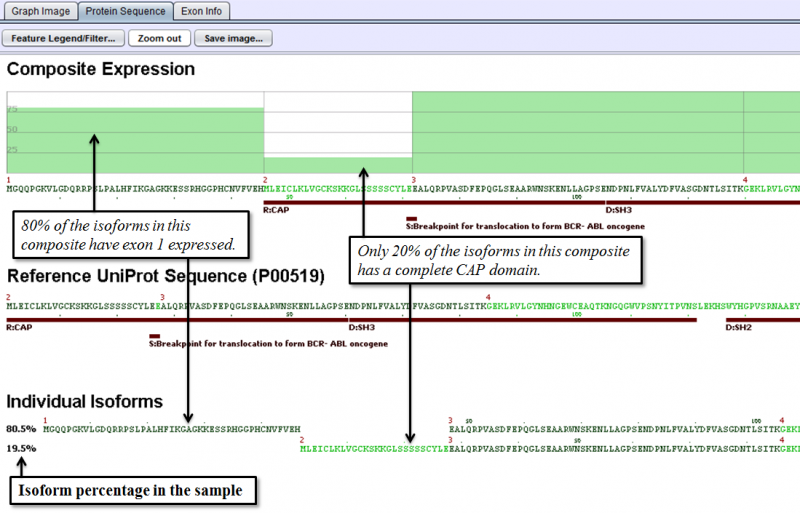
Exon Info Tab of the Detail Panel shows the location, properties and nucleotide sequences of each exon in the gene model. More information can be found in the GUI Overview section.