SpliceSeq
| Overview | |
| Description | A tool for investigating alternative mRNA splicing in next generation mRNA sequence data |
| Development Information | |
| Language | Java |
| Current version | 2.1 |
| Platforms | Platform independent |
| License | Freely available for academic and commercial use |
| Status | Active |
| Last updated | February 21, 2014 |
| References | |
| Citation | Ryan, M. C., Cleland, J., Kim, R.G., Wong, W. C., Weinstein, J. N., SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts, Bioinformatics 28 (18) p.2385 (2012). https://doi.org/10.1093/bioinformatics/bts452 |
| Help and Support | |
| Contact | MDACC-Bioinfo-IT-Admin@mdanderson.org |
SpliceSeq
2/21/2014 - A patch release, SpliceSeq 2.1 has been released. The patch includes minor bug fixes, and update of the SpliceGraph database and turns off a component in group / group comparison analysis that was causing excessively long loads.
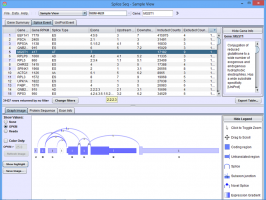
SpliceViewer is a Java application that allows researchers to investigate alternative mRNA splicing patterns in data from high-throughput mRNA sequencing studies. Sequence reads are mapped to splice graphs that unambiguously quantify the inclusion level of each exon and splice junction. The graphs are then traversed to predict the protein isoforms that are likely to result from the observed exon and splice junction reads. UniProt annotations are mapped to each protein isoform to identify potential functional impacts of alternative splicing.
This tool may be used on a single RNASeq sample to identify genes with multiple spliceforms, on a pair of samples to identify differential splicing between the two, or on groups of samples to identify statistically significant group level differences in splicing patterns.
SpliceSeq can be installed locally to analyze your own mRNA-Seq data.
Tutorials
Several YouTube tutorials have been developed for SpliceSeq. Please maximize and switch to 1080p resolution for best viewing.
Documentation
Click on the images below for basic help on installation, usage, and methodology.

|

|
|
|
|
|
|

|
|
If you have further questions, please email us at
binf@insilico.us.com
.
Release History
2/21/2014 - Splice 2.1
Version 2.1 includes a modification to the SGAnalyzePrograms.properties file to remove running of the ComparePatch, GroupPatch, and CompareGroupPatch modules during data analysis/load. These components were causing very long load times on large studies. These components have a very minor effect on differential splicing analysis. New versions are in development. The release also includes an updated SpliceGraph database and minor adjustments to the Alternate Promoter / Alternate Terminator event detection to reduce internal exons being identified as AP/AT events. The 2.1 version of SpliceSeq can be used without a database upgrade.
8/13/2013 - Splice 2.0
Version 2.0 uses Percent Splice In (PSI) for differential splicing analysis. It has splice event lists with p-values that can be filtered / sorted by the users. It also includes novel splice detection and improved splice event detection algorithms. The user interface has a new look and feel and includes many usability improvements and a composite protein impact view.
1/31/2013 - SpliceSeq 1.2.1
Fixes made to correct issues with grayed out Load Study Data menu item, classpath issues, arff file syntax checks, display of log/stat info, and grayed out Run buttons. No data changes from 1.2 version. Patch may be applied without re-loading sample data.
1/7/2013 - SpliceSeq 1.2
Version 1.2 includes a much easier study data processing flow, enhanced splice detection, bug fixes, and a gene information display window.
5/1/2012 - SpliceSeq 1.1
Version 1.1 includes sample annotation, sample groups, and group comparison.
12/21/2011 - SpliceSeq 1.0 Released for validation / testing.